`polySimIBD`: A Simple Malaria Genetic Model
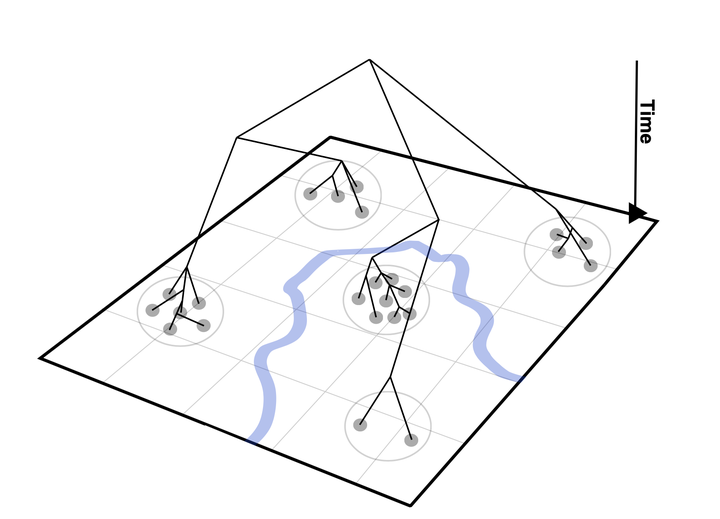
The goal of polySimIBD is to perform forwards in-time simulation of malaria population genetics. The model uses a discrete-time, discrete-loci structured Wright Fisher approximation to account for (simplified) malaria transmission dynamics and can be parameterized across demes to include a spatial component.
The model was originally used in Verity & Aydemir & Brazeau et al. 2020 in Nature Communications1 to assess the bias in our identity by descent calculation.
Verity R, Aydemir O, Brazeau NF, Watson OJ, Hathaway NJ, Mwandagalirwa MK, Marsh PW, Thwai K, Fulton T, Denton M, Morgan AP, Parr JB, Tumwebaze PK, Conrad M, Rosenthal PJ, Ishengoma DS, Ngondi J, Gutman J, Mulenga M, Norris DE, Moss WJ, Mensah BA, Myers-Hansen JL, Ghansah A, Tshefu AK, Ghani AC, Meshnick SR, Bailey JA, Juliano JJ. The impact of antimalarial resistance on the genetic structure of Plasmodium falciparum in the DRC. Nat Commun. 2020 Apr 30;11(1):2107. doi: 10.1038/s41467-020-15779-8. PMID: 32355199; PMCID: PMC7192906. ↩︎