Antimalarial Resistance in the DRC
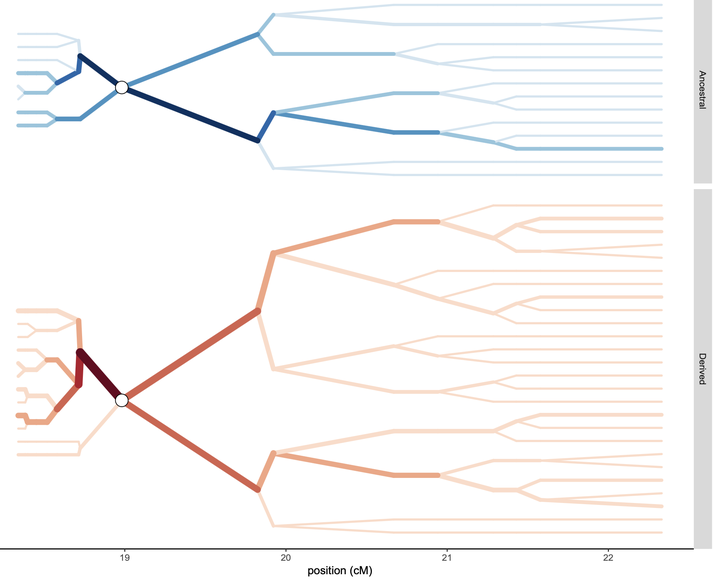
Using a massively parallel amplicon sequencing barcode approach, we used genomic analyses to demonstrate that the DRC appears to be a “mixing pot” for P. falciparum infections across Sub-Saharan Africa. In addition, we identified strong signals of selection to antimalarial drugs in the DRC that likely have driven gene flow patterns across the country. Alongside analytic support, I lead the analyses on detecting selection using adapted extended haplotype heterozygosity approaches. In addition, I led the bioinformatic analysis identifying the variants for the aforementioned barcode used in the study. This barcode has been used extensively by our research group, as well as several domestic and international groups (initial publication: Verity/Aydemir/Brazeau et al. 2020, Nat Comms).