Research & Interests
Below, I briefly summarize my research to date and elaborate on my interests and what I hope to continue to work on in the future. At the end of this page, you will find current and past projects.
Infectious Disease Modeling
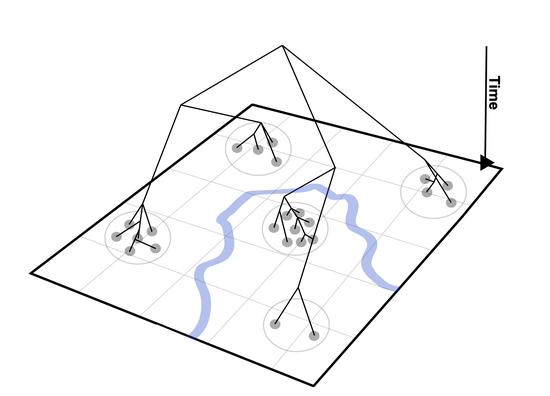
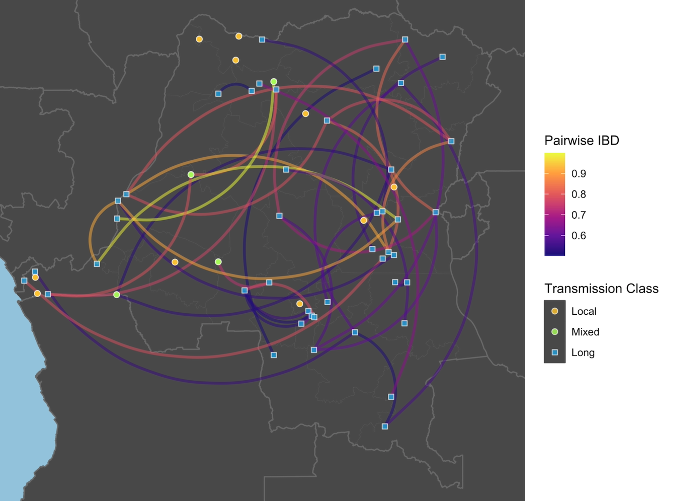
My infectious disease modeling work spans statistical, spatial, evolutionary, and computational approaches. In one framework, I examined who became infected, where infections occurred, and the origin of pathogens in regions previously considered resilient to transmission. To evaluate who was at risk, I applied causal inference methods using inverse probability treatment weights generated through the SuperLearner ensemble. To determine where infections clustered, I used Bayesian Gaussian spatial process models. To assess origin, I generated and analyzed pathogen genetic data.
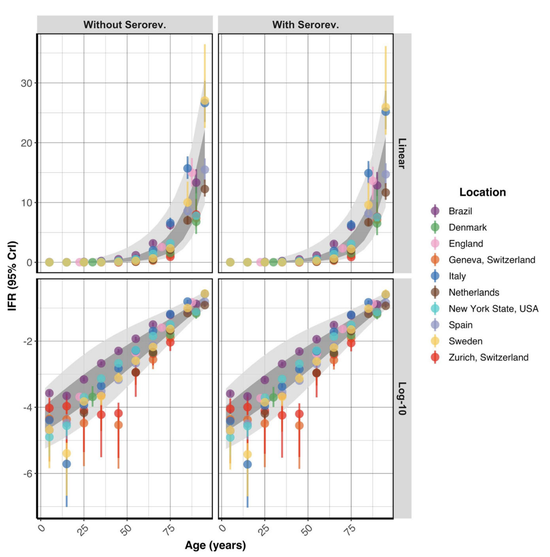
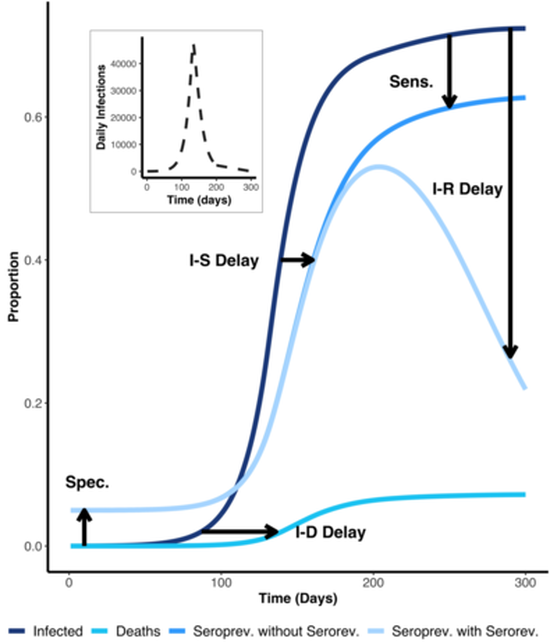
For research questions involving severity or burden estimation, I have implemented Bayesian statistical models and contributed to mathematical modeling for scenario planning. Most recently, I have explored how structural features of contact networks hinder pandemic prediction and evaluated AI/ML-driven agent-based models for improving forecasting.
Broadly, I am interested in the intersections of statistical modeling, infectious disease dynamics, machine learning/AI, and spatial analysis. I aim to use these tools to understand transmission and characterize antimicrobial resistance evolution and ultimately identify points where within-host and between-host propagation can be disrupted.

Bioinformatics, Genomics, Statistical Genetics
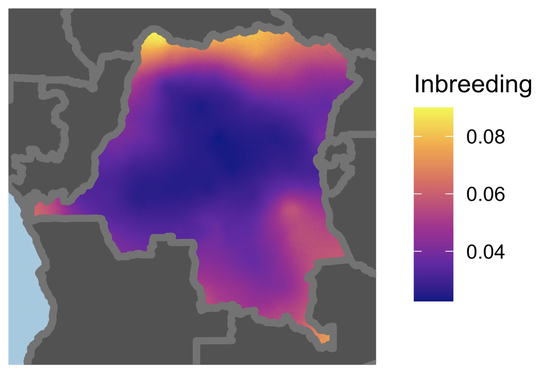
Molecular surveillance and pathogen genomics critically inform infectious disease epidemiology and intervention planning. Much of my work focuses on antimicrobial resistance, especially putative antimalarial resistance. I have used genome-wide barcodes to study how antimalarial selection pressures shape Plasmodium falciparum population structure in the Democratic Republic of the Congo and performed whole-genome sequencing analyses to identify novel antimicrobial resistance mutations in Staphylococcus epidermidis.
I also develop statistical genetics methods, including:
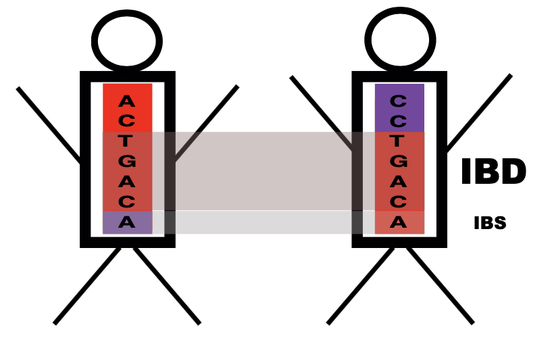
- Estimating local inbreeding with
DISCent, relevant for sink–source dynamics and public health intervention design. - Simulating malaria transmission dynamics with
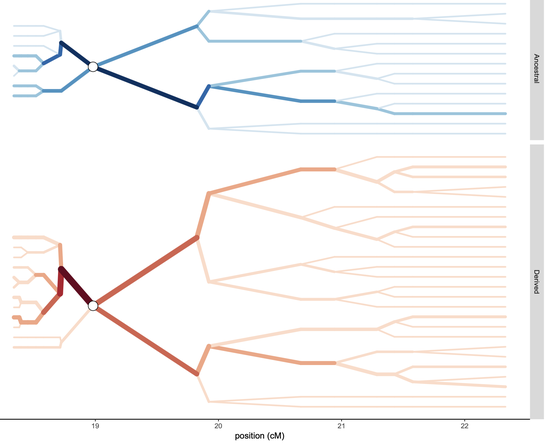
polySimIBDusing a forwards in-time simulation with a spatial discrete-time, discrete-loci structured Wright Fisher model. - Inferring identity-by-descent in polyclonal malaria infections with
HMMERTIME.
Global Health & Medicine, Translational Research
Global medicine has become local medicine— and vice versa. I am committed to supporting capacity building and collaborative research in low- and middle-income countries and historically underserved communities. My work aims to integrate computational, molecular, and clinical perspectives to improve infectious disease prevention, diagnosis, and treatment across diverse settings.